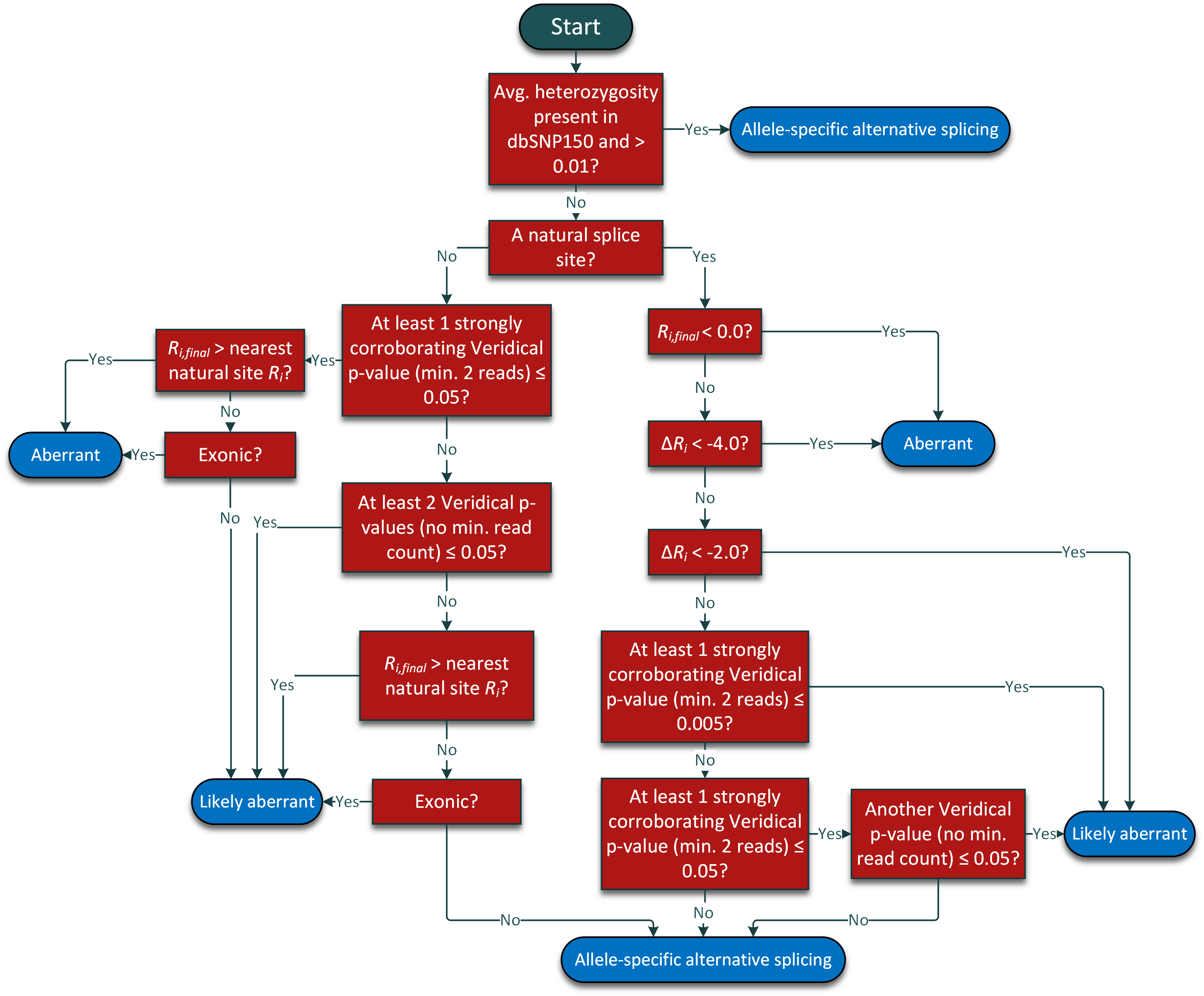
cases - click to expand
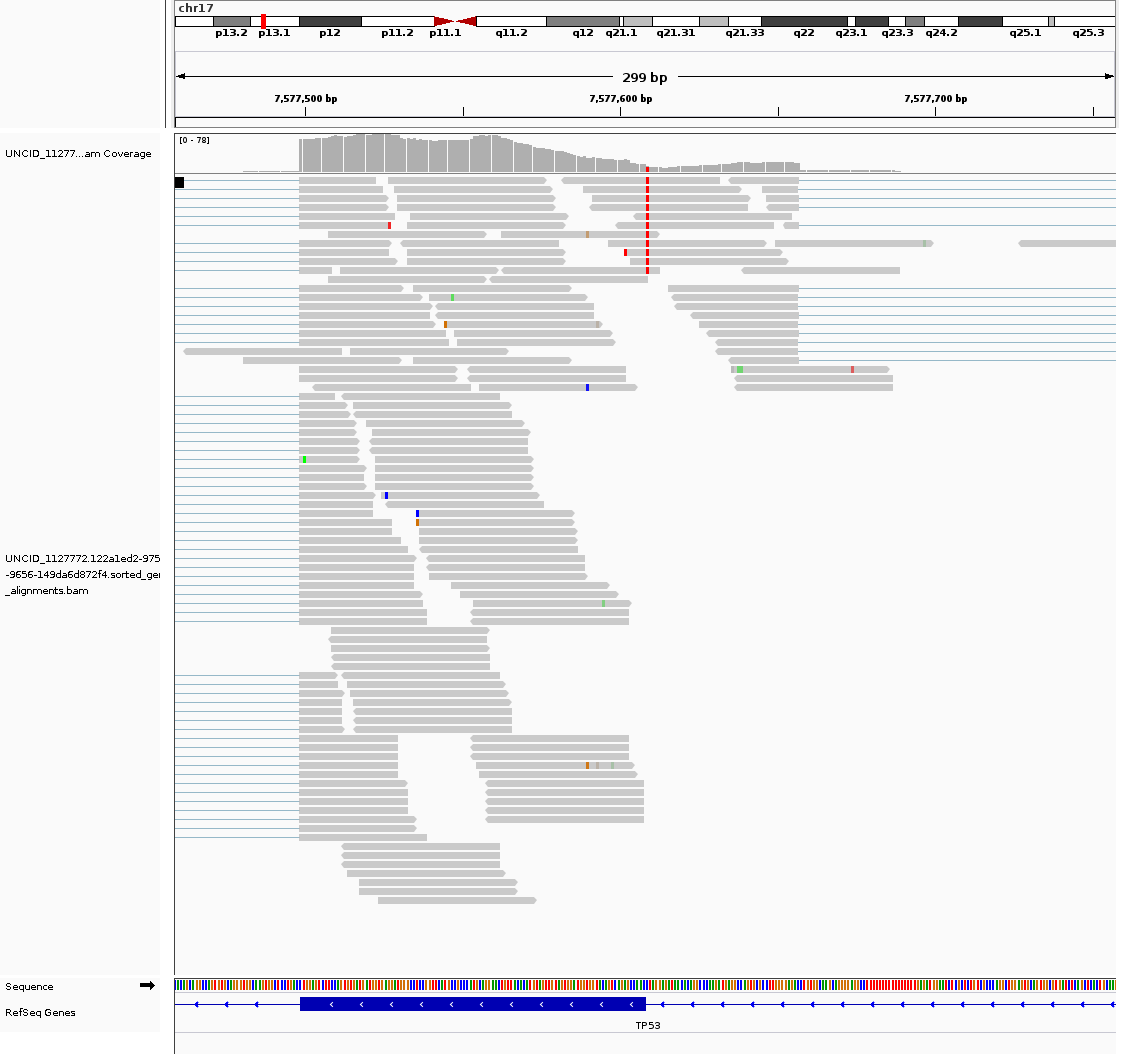
allele-specific alternative splicing
likely aberrant
View TCGA-A2-A1G1 metadata (NCI Genomic Data Commons)
Veridical validated this mutation based on 12 reads which span the splice junction and extend into the intron.
Veridical validated this mutation based on 11 reads which span the splice junction, extend into the intron, and contain the mutation in question.
| Evidence Type | Cryptic | Anti-Cryptic | Exon Skipping | Intron Inclusion | Intron Inclusion with Mutation |
|---|---|---|---|---|---|
| Junction spanning | 0 | 0 | 0 | 12 (p=0.0002) | 11 (p=0) |
| Read Abundance | 0 | 0 | 0 | 29 (p=0.1813) | 0 |
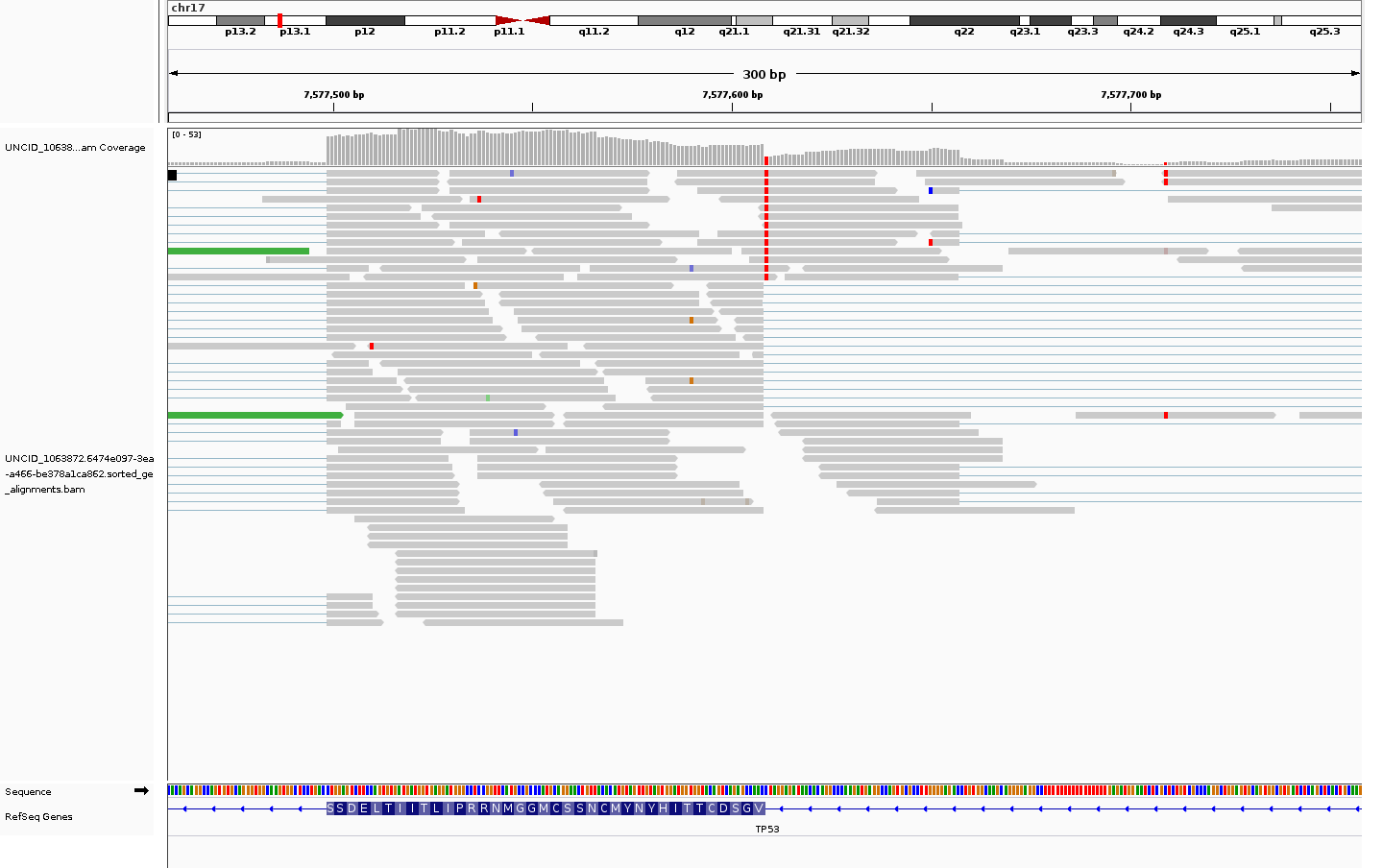
allele-specific alternative splicing
likely aberrant
View TCGA-CV-7104 metadata (NCI Genomic Data Commons)
Veridical validated this mutation based on 1 reads which span the splice junction, extend into the intron, and contain the mutation in question.
| Evidence Type | Cryptic | Anti-Cryptic | Exon Skipping | Intron Inclusion | Intron Inclusion with Mutation |
|---|---|---|---|---|---|
| Junction spanning | 0 | 0 | 0 | 4 (p=0.2803) | 1 (p=0) |
| Read Abundance | 0 | 0 | 0 | 4 (p=0.8082) | 0 |
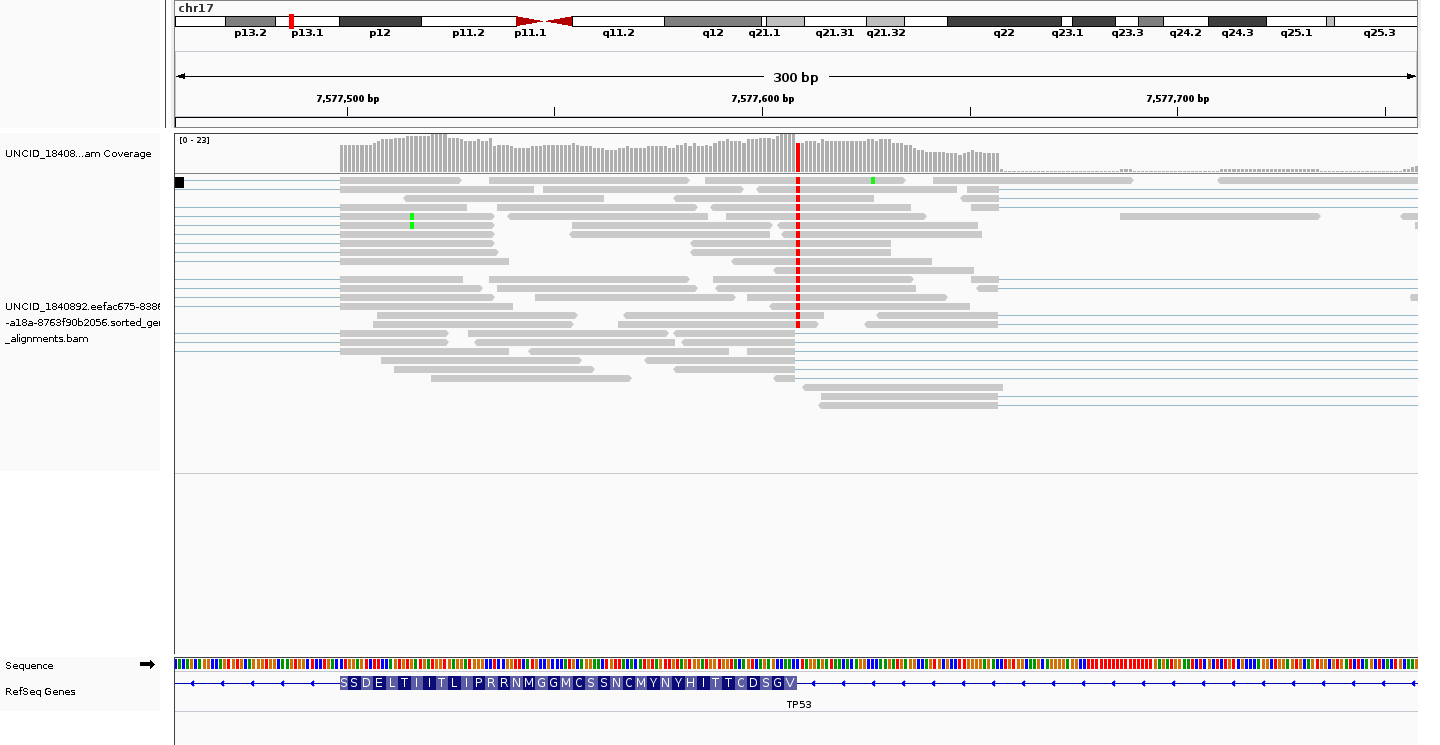
allele-specific alternative splicing
likely aberrant
View TCGA-77-6844 metadata (NCI Genomic Data Commons)
Veridical validated this mutation based on 12 reads which span the splice junction and extend into the intron.
Veridical validated this mutation based on 12 reads which span the splice junction, extend into the intron, and contain the mutation in question.
Veridical validated this mutation based on 85 reads each of which either overlap the splice boundary or are wholly contained within an intron.
| Evidence Type | Cryptic | Anti-Cryptic | Exon Skipping | Intron Inclusion | Intron Inclusion with Mutation |
|---|---|---|---|---|---|
| Junction spanning | 0 | 0 | 0 | 12 (p=0.0064) | 12 (p=0) |
| Read Abundance | 0 | 0 | 0 | 85 (p=0.0001) | 0 |
allele-specific alternative splicing
likely aberrant
View TCGA-52-7810 metadata (NCI Genomic Data Commons)
Veridical validated this mutation based on 17 reads which span the splice junction and extend into the intron.
Veridical validated this mutation based on 17 reads which span the splice junction, extend into the intron, and contain the mutation in question.
| Evidence Type | Cryptic | Anti-Cryptic | Exon Skipping | Intron Inclusion | Intron Inclusion with Mutation |
|---|---|---|---|---|---|
| Junction spanning | 0 | 0 | 0 | 17 (p=0.0004) | 17 (p=0) |
| Read Abundance | 0 | 0 | 0 | 26 (p=0.1893) | 0 |
allele-specific alternative splicing
likely aberrant
View TCGA-KO-8404 metadata (NCI Genomic Data Commons)
Veridical validated this mutation based on 2 reads which span the splice junction, extend into the intron, and contain the mutation in question.
Veridical validated this mutation based on 26 reads each of which either overlap the splice boundary or are wholly contained within an intron.
| Evidence Type | Cryptic | Anti-Cryptic | Exon Skipping | Intron Inclusion | Intron Inclusion with Mutation |
|---|---|---|---|---|---|
| Junction spanning | 0 | 0 | 0 | 2 (p=0.1587) | 2 (p=0) |
| Read Abundance | 0 | 0 | 0 | 26 (p=0) | 0 |
allele-specific alternative splicing
likely aberrant
View TCGA-04-1362 metadata (NCI Genomic Data Commons)
Veridical validated this mutation based on 35 reads which span the splice junction, extend into the intron, and contain the mutation in question.
Veridical validated this mutation based on 125 reads each of which either overlap the splice boundary or are wholly contained within an intron.
| Evidence Type | Cryptic | Anti-Cryptic | Exon Skipping | Intron Inclusion | Intron Inclusion with Mutation |
|---|---|---|---|---|---|
| Junction spanning | 0 | 0 | 0 | 38 (p=0.4624) | 35 (p=0) |
| Read Abundance | 0 | 0 | 0 | 125 (p=0.0009) | 0 |